Forecasting Whole-Brain Connectomics – A Kurzweilian Approach – Article by Dan Elton

Daniel C. Elton, Ph.D.
Editor’s Note: In this article, U.S. Transhumanist Party Director of Scholarship Dr. Daniel C. Elton describes the recent advances in mapping the connectomes of various organisms, as well as the technological advances that would be needed to achieve effective human whole-brain emulation. Given extensive discussion of these subjects among U.S. Transhumanist Party members, including at the Virtual Enlightenment Salon of September 27, 2020, with Kenneth Hayworth and Robert McIntyre, it is fitting for the U.S. Transhumanist Party to feature this systematic exploration by Dr. Elton into what has been achieved in the field of connectomics already and what it would practically take for human whole-brain emulation to become a reality. As Dr. Elton convincingly illustrates, this possibility is still several decades away, but some steady progress has been made in recent years as well.
~ Gennady Stolyarov II, Chairman, United States Transhumanist Party, March 7, 2021
The connectome of an organism is a map of all neurons and their connections. This may be thought of as a graph with the neurons as nodes and synaptic connections as edges. Here we take the term ‘connectome’ to refer to the graph and the underlying electron microscopy images of the neurons, which contain much more information. However, to successfully simulate an organism’s brain using a connectome, more information will be needed. Retrieving a detailed scan of an entire brain and mapping all the neurons is a prerequisite for whole-brain emulation. In their landmark 2008 paper, “Whole Brain Emulation: A Roadmap“, transhumanists Anders Sandberg and Nick Bostrom construct a detailed “technology tree” showing the prerequisite technologies for realizing whole brain emulation:

In this article, we focus on the “scanning” component along with part of the “translation” component, namely neuronal tracing. By plotting technological progress on a logarithmic plot, similar to how Kurzweil does, we attempt to forecast how many decades away we are from being able to scan an entire human brain (and trace/segment all neurons to determine the connectome). Of course, while Kurzweilian projections have been known to hold (most famously for Moore’s law), we caution that the start of a logistic function can look like an exponential function. In other words, exponential trends can and often do plateau. As any investment advisor would say, “past returns are no guarantee of future results”.
The complete connectome of the nematode worm (Caenorhabditis Elegans) was published in 1986. A complete set of images of the fruit fly (Drosophila melanogaster) was published in 2018. However, all of the neurons and their connections have not yet been segmented or traced. In January 2020 researchers published the connectome of the central brain of the fruit fly, containing 25,000 neurons, which to my knowledge is the largest connectomics dataset published to date.
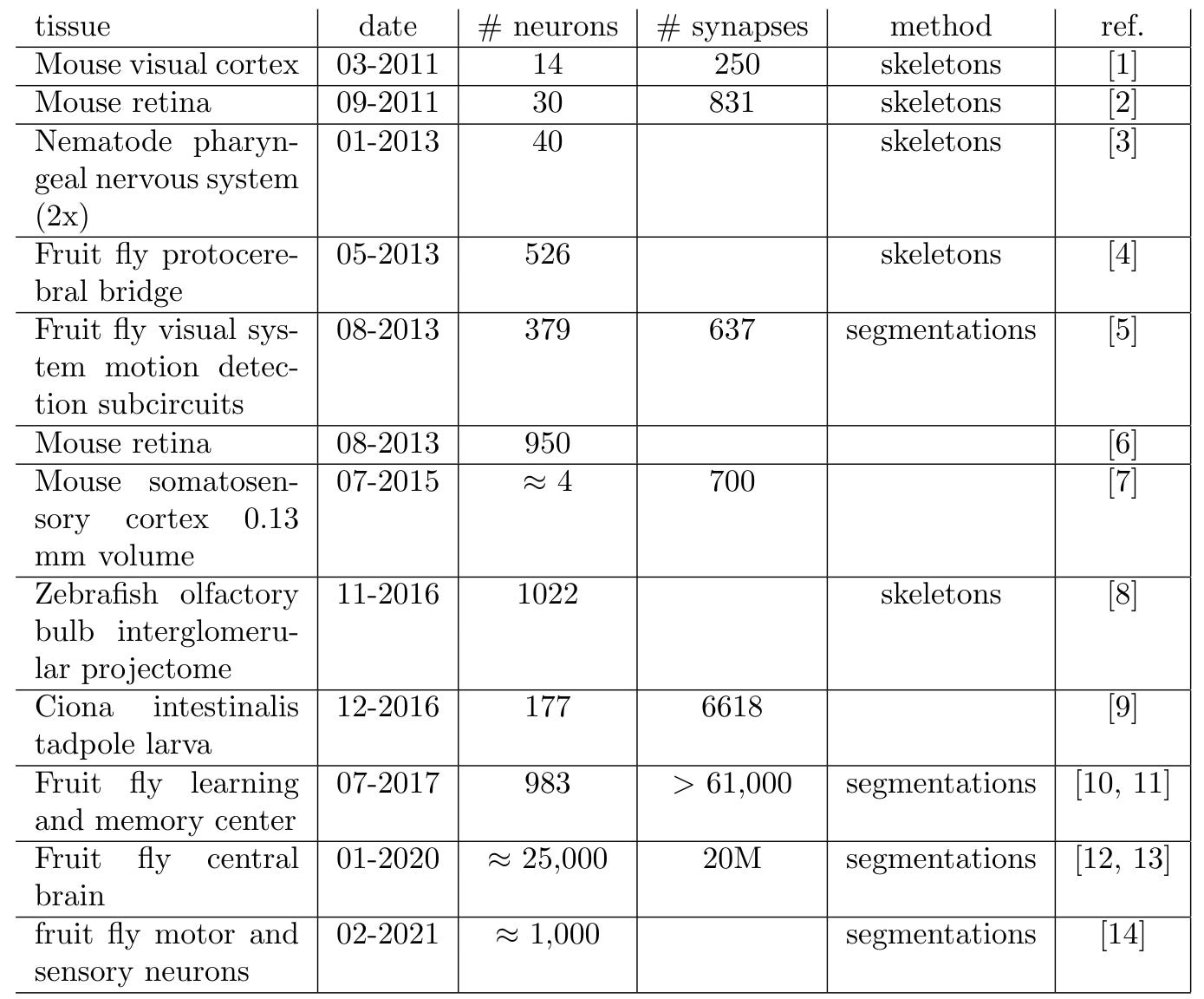
I thought it would be fun/interesting to plot the progress of connectomics over time and try to extrapolate out any trend observed. So, I did a literature search for all studies to date which either traced or segmented neurons and marked out synapses in electron microscopy data:
[1] D. D. Bock, et al. “Network anatomy and in vivo physiology of visual cortical neurons”, Nature 471 (7337) (2011) 177–182. doi:10.1038/nature09802. [link]
[2] K. L. Briggman, M. Helmstaedter, W. Denk, Wiring specificity in the direction-selectivity circuit of the retina, Nature 471 (7337) (2011) 183–188. [link]
[3] D. J. Bumbarger, M. Riebesell, C. Rodelsperger, R. J. Sommer, System-wide rewiring underlies behavioral differences in predatory and bacterial-feeding nematodes, Cell 152 (1-2) (2013) 109–119. [link]
[4] C.-Y. Lin, et al., A comprehensive wiring diagram of the protocerebral bridge for visual information processing in the drosophila brain, Cell Reports 3 (5) (2013) 1739–1753. [link]
[5] S. ya Takemura, et al., A visual motion detection circuit suggested by drosophila connectomics, Nature 500 (7461) (2013) 175–181. [link]
[6] M. Helmstaedter, K. L. Briggman, S. C. Turaga, V. Jain, H. S. Seung, W. Denk, Connectomic reconstruction of the inner plexiform layer in the mouse retina, Nature 500 (7461) (2013) 168–174. [link]
[7] N. Kasthuri, et al., Saturated reconstruction of a volume of neocortex, Cell 162 (3) (2015) 648–661. [link]
[8] A. A. Wanner et al., 3-dimensional electron microscopic imaging of the zebrafish olfactory bulb and dense reconstruction of neurons, Scientific Data 3 (1). [link]
[9] K. Ryan, Z. Lu, I. A. Meinertzhagen, The CNS connectome of a tadpole larva of Ciona intestinalis (l.) highlights sidedness in the brain of a chordate sibling, eLife 5 (2016) [link]
[10] S.-y. Takemura, et al., A connectome of a learning and memory center in the adult Drosophila brain, eLife 6 (2017). [link]
[11] K. Eichler, et al., The complete connectome of a learning and memory centre in an insect brain, Nature 548 (7666) (2017) 175–182. [link]
[12] C. S. Xu, et al., A connectome of the adult drosophila central brain (preprint) [link]
[13] L. K. Scheffer, et al., A connectome and analysis of the adult drosophila central brain, eLife 9 (2020). [link]
[14] J. S. Phelps, et al., Reconstruction of motor control circuits in adult drosophila using automated transmission electron microscopy, Cell 184 (3) (2021) 759–774.e18. [link]
Next I plotted most of the data for the number of neurons versus the date of publication:
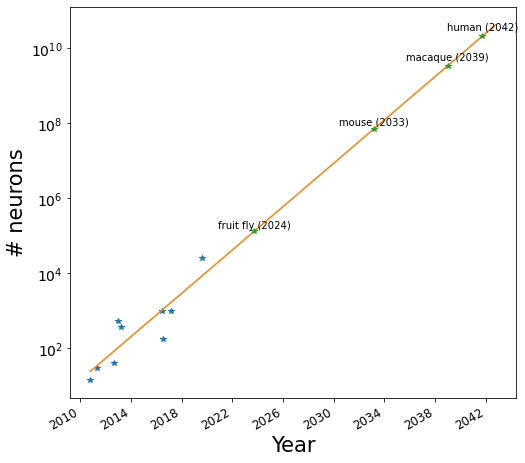
Next I did linear regression on the (year, log(# neurons)) data which is equivalent to fitting an exponential function to the data. (The reason for fitting the data in this way was to avoid the bias that occurs when fitting an exponential function with least-squares regression that leads to the larger values on the y axis being fit more accurately than smaller ones.) After doing the linear regression I extrapolated it forward in time.
The projection for the fruit-fly connectome (2024) seems about right. If anything, we may see it slightly sooner. It will be interesting to see how much longer it will take before we have physically realistic models of the fruit fly and fruit-fly behavior. U.S. Transhumanist Party member Logan T. Collins has advocated for building biophysically and behaviorally realistic models of insects to better understand nervous systems. For one thing, interesting neuroscience experiments may be performed on a simulated “virtual fly” much faster and easier than on a real fly (for instance, certain neurons may be removed or manipulated, and the effects on the virtual fly’s behavior observed). A project to produce the mouse brain connectome is underway, and again, the date extrapolated to — 2033 — seems plausible if the funding for the project continues. Beyond that though, I have very little idea how plausible the projections are!
Here are some numbers that show the challenges just with scanning the entire brain (not to mention segmenting/tracing all the neurons accurately!).
Assuming an isotropic voxel size of 20 nm, it is estimated that storing the images of an entire human brain would require 175 exabytes of storage. It seems we are approaching hard drives which cost about 1.5 cents per gigabyte. Even at those exorbitantly low prices, it would still cost $2.6 billion to store all those images!
The volume of the human brain is about 1.2 x 10^6 cubic millimeters. The Zeiss MultiSEM contains either 61 or even 91 electron beams which scan a sample in parallel. According to a Zeiss video presentation from April 8th, 2020, it can scan a 1×1 mm area at 4 nm resolution in 6.5 minutes. Assuming a slice thickness of 20 nm, a single such machine would require 742,009 years to scan the entire brain!
X-ray holographic nano-tomography might be the path forward …
Dan Elton, Ph. D., is Director of Scholarship for the U.S. Transhumanist Party. You can find him on Twitter at @moreisdifferent, where he accepts direct messages. If you like his content, check out his website and subscribe to his newsletter on Substack.

One thought on “Forecasting Whole-Brain Connectomics – A Kurzweilian Approach – Article by Dan Elton”
That is an insane amount of time to be able to scan the brain with the Zeiss.
Thanks for introducing the concept of holographic tomography, would be great to skim the full paper.